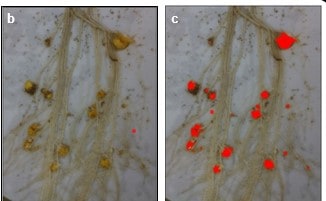
Our study proposes a rapid, accurate and low-cost phenotyping tool for the sunflower/O. cumana interaction under controlled conditions through image analysis for broomrape tubercle analysis at early stages of the interaction. We optimized the phenotyping of sunflower/O. cumana interactions by using rhizotrons (transparent Plexiglas boxes) in a growth chamber to control culture conditions and Orobanche inoculum. We used a Raspberry Pi computer with a picamera for acquiring images of inoculated sunflower roots 3 weeks post inoculation. We set up a macro using ImageJ free software for the automatic counting of the number of tubercles. This phenotyping tool was named RhizOSun. We evaluated five sunflower genotypes inoculated with two O. cumana races and showed that automatic counting of the number of tubercles using RhizOSun was highly correlated with manual time-consuming counting and could be efficiently used for screening sunflower genotypes at the tubercle stage.