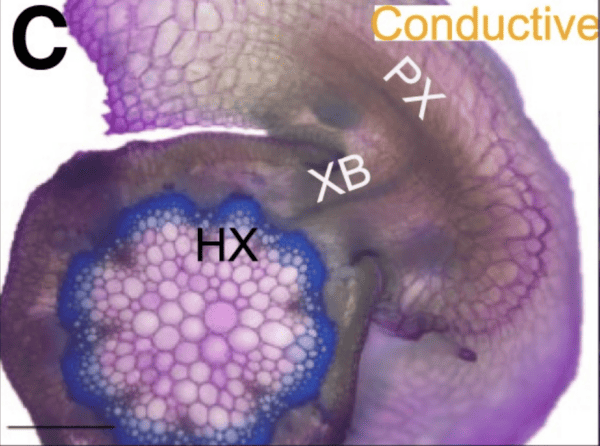
Cuscuta campestris produces many unique microRNAs only at the haustorial interface with hosts. Our previously published work showed that these interface-induced microRNAs can target host messenger RNAs (mRNAs). The targeted host mRNAs generally function in immune responses, vascular system functions, or hormone signal transduction. Our overall hypothesis is that the Cuscuta interface-induced microRNAs act in a trans-species manner to suppress certain host proteins, and in doing so provide an advantage to the parasite. In this study by Hudzik et al. (https://doi.org/10.1093/plcell/koad076) we examined several aspects of these microRNAs. We found that the induction of the C. campestris interface-induced microRNAs was host-independent: Induction could be equally stimulated from haustoria that were induced to form in the absence of any plant hosts. This shows that there is no host-selectivity in the deployment of these microRNAs. We also analyzed the microRNA genes in the C. campestris genome that give rise to the interface-induced microRNAs. We found to our surprise that the interface-induced microRNA genes shared a common cis-regulatory motif in their promoters. This motif is identical in sequence and spacing to the upstream sequence element that has long been known to drive the RNA polymerase III-catalyzed transcription of small nuclear RNAs (snRNAs) in plants. We demonstrated that this cis-element is required for interface-induced microRNA accumulation. We also demonstrated that RNA polymerase III does indeed transcribe the C. campestris interface-induced microRNA genes. This is a surprising result since all other “canonical” microRNA genes, both in C. campestris and all other plants, are transcribed by RNA polymerase II. We speculate that transcription by RNA pol III somehow enhances the export of the C. campestris interface-induced microRNAs from parasite tissues and into host tissues.