Strigolactones (SLs) were initially discovered as germination stimulants that induce the seed germination of parasitic plants of the Orobanchaceae (Striga, Alectra, Phelipanche and Orobanche genera). Half a century after this intriguing biological discovery, it was demonstrated that SLs are actually beneficial for plants, as they induce hyphal branching in arbuscular mycorrhizal (AM) fungi. Another 3 yr later, SLs were revealed to be a class of plant hormones regulating plant architecture and development.
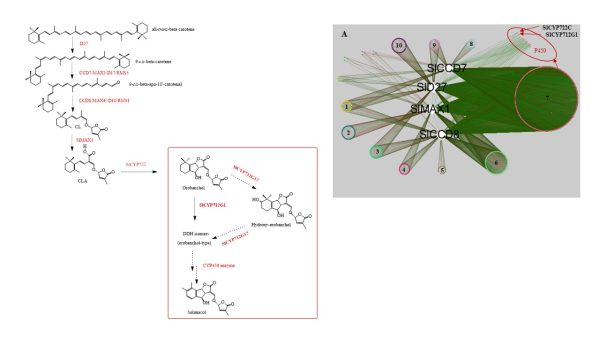
The biosynthetic pathway of SLs in tomato has been partially elucidated, but the structural diversity in tomato SLs predicts that additional biosynthetic steps are required. Here, root RNA-seq data and co-expression analysis were used for SL biosynthetic gene discovery. This strategy resulted in a candidate gene list containing several cytochrome P450s. Heterologous expression in Nicotiana benthamiana and yeast showed that one of these, CYP712G1, can catalyse the double oxidation of orobanchol, resulting in the formation of three didehydro-orobanchol (DDH) isomers. Virus-induced gene silencing and heterologous expression in yeast showed that one of these DDH isomers is converted to solanacol, one of the most abundant SLs in tomato root exudate. Protein modelling and substrate docking analysis suggest that hydroxy-orbanchol is the likely intermediate in the conversion from orobanchol to the DDH isomers. Phylogenetic analysis demonstrated the occurrence of CYP712G1 homologues in the Eudicots only, which fits with the reports on DDH isomers in that clade. Protein modelling and orobanchol docking of the putative tobacco CYP712G1 homologue suggest that it can convert orobanchol to similar DDH isomers as tomato.
See: https://nph.onlinelibrary.wiley.com/doi/full/10.1111/nph.18272